Angelique D’Hont, France Denoeud, Jean-Marc Aury Franc-Christophe Baurens, Françoise Carreel, Olivier Garsmeur, Benjamin Noel, Stephanie Bocs, Gaetan Droc, Mathieu Rouard, Corinne Da Silva Kamel Jabbari, Celine Cardi, Julie Poulain, Marlene Souquet, Karine Labadie, Cyril Jourda, Juliette Lengelle, Marguerite Rodier-Goud, Adriana Alberti, Maria Bernard, Margot Correa, Saravanaraj Ayyampalayam, Michael R. Mckain, Jim Leebens-Mack, Diane Burgess, Mike Freeling, Didier Mbeguie-A-Mbeguie, Matthieu Chabannes, Thomas Wicker, Olivier Panaud, Jose Barbosa, Eva Hribova, Pat Heslop-Harrison, Remy Habas, Ronan Rivallan, Philippe Francois, Claire Poiron, Andrzej Kilian, Dheema Burthia, Christophe Jenny, Frederic Bakry, Spencer Brown, Valentin Guignon, Gert Kema, Miguel Dita, Cees Waalwijk, Steeve Joseph, Anne Dievart, Olivier Jaillon, Julie Leclercq, Xavier Argout, Eric Lyons, Ana Almeida, Mouna Jeridi, Jaroslav Dolezel, Nicolas Roux, Ange-Marie Risterucci, Jean Weissenbach, Manuel Ruiz, Jean-Christophe Glaszmann, Francis Quetier, Nabila Yahiaoui & Patrick Wincker, "The Banana (Musa acuminata) genome and the evolution of monocotyledonous plants," Nature, 2012
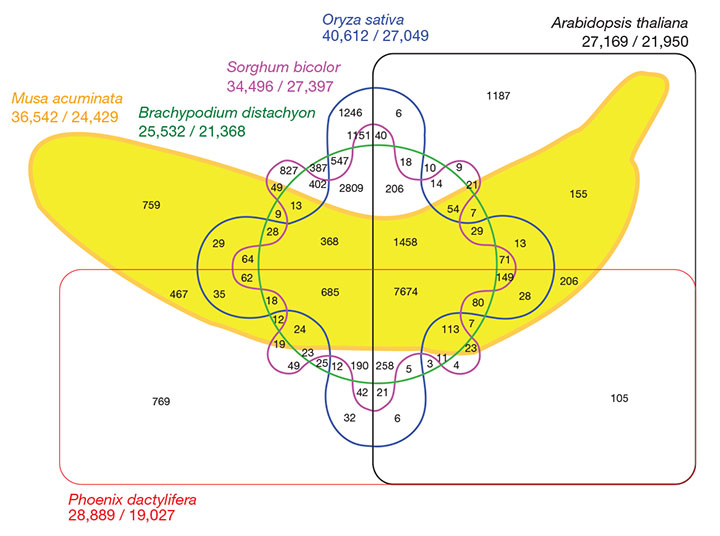
SIX-WAY VENN DIAGRAM SHOWING THE DISTRIBUTION OF SHARED GENE FAMILIES (SEQUENCE CLUSTERS) AMONG M. ACUMINATA, P. DACTYLIFERA, ARABIDOPSIS THALIANA, ORYZA SATIVA, SORGHUM BICOLOR AND BRACHYPODIUM DISTACHYON GENOMES
Suggested by Darren Story
Interestingly, many specific clusters (809 in our setting) proved specific to Poaceae, suggesting a high level of gene divergence and diversification within the grass lineage. Specific Musa clusters (759) were enriched in genes encoding transcription factors (for example, Myb and AP2/ERF families), defence-related proteins, enzymes of cell-wall biosynthesis and enzymes of secondary metabolism.
![[ToC]](14_6_toc_t_off.jpg)